Jed Quiaoit
Samantha Himegarner
AP Biology 🧬
358 resourcesSee Units
Regulatory Sequences
Regulatory sequences are regions of DNA that play a crucial role in controlling gene expression in terms of increasing or decreasing the expression of certain genes in the organism; essentially, they can turn a gene "on" or "off." These sequences are often located near or within the promoter regions of genes, and they interact with specific regulatory proteins to either activate or repress transcription.

Source: ResearchGate
There are several different types of regulatory sequences, each with their own unique functions. For example, enhancers are sequences that can increase the level of transcription of a gene, while silencers can decrease it. Promoters are sequences that provide the binding site for the RNA polymerase and other initiation factors, while terminators are sequences that signal the end of transcription.
Regulatory proteins, also known as transcription factors, bind to specific regulatory sequences and control the rate of transcription. These proteins can either activate or repress transcription by recruiting or inhibiting the binding of the RNA polymerase to the promoter region. ⬆️
Regulatory sequences and proteins work together to ensure that genes are expressed at the appropriate time and level in different cells and tissues. Dysregulation of these sequences and proteins can lead to various diseases, including cancer. Therefore, understanding the function and mechanism of regulatory sequences and proteins is crucial for the development of new therapeutic strategies for diseases.
Epigenetic Changes
Epigenetics refers to the study of heritable changes in gene function that occur without changes to the underlying DNA sequence. These changes can affect the expression of genes by influencing the accessibility of the DNA to the transcription machinery. Epigenetic modifications can occur on both DNA and histones, which are the proteins around which DNA is wrapped to form chromosomes. ⚽
One of the most well-known epigenetic modifications is methylation of cytosine bases in DNA, which can lead to the repression of gene expression by recruiting methyl-DNA binding proteins. Another common epigenetic modification is acetylation and methylation of histones, which can change the structure of chromatin and affect the accessibility of the DNA to the transcription machinery.
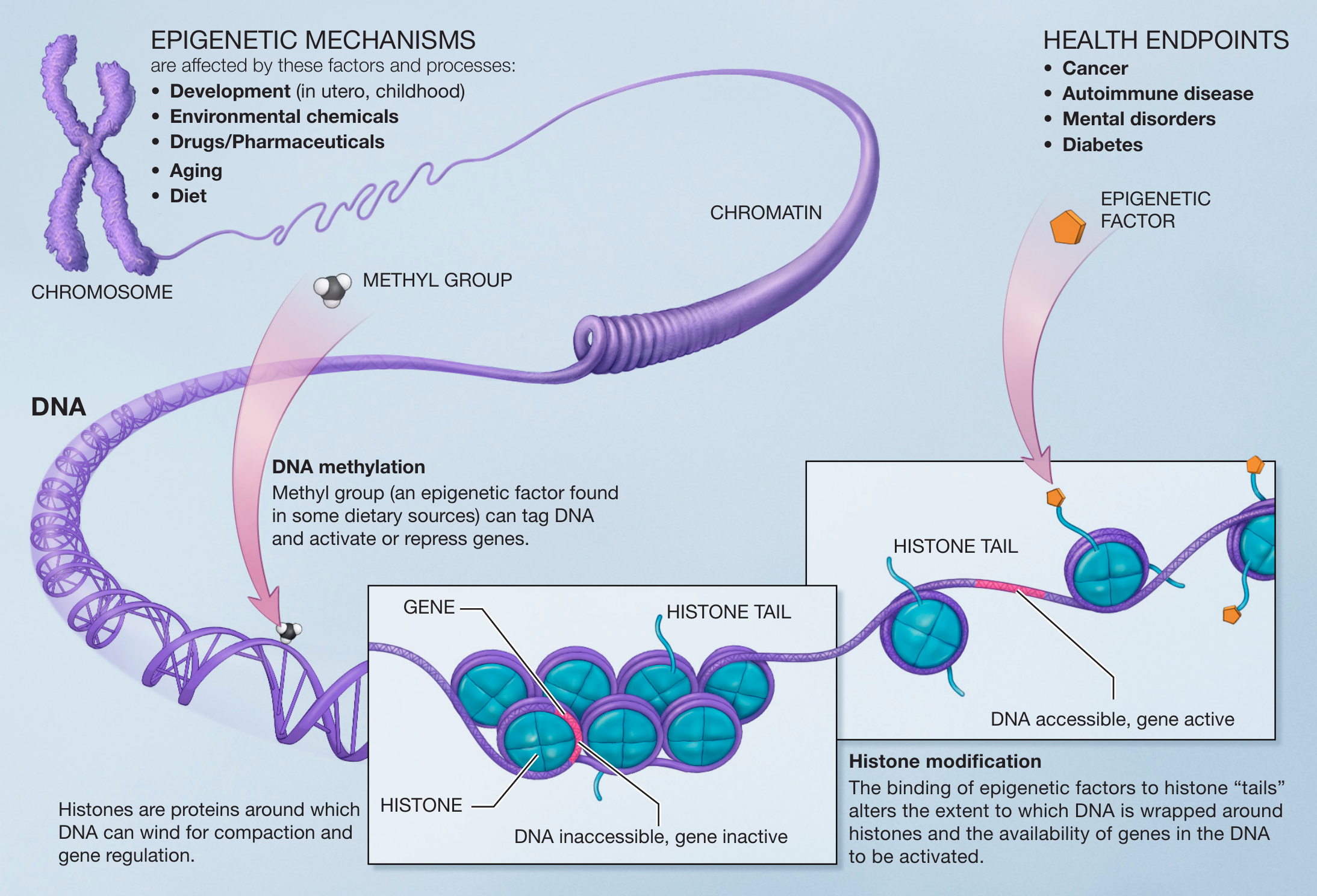
Source: Wikimedia Commons
Epigenetic modifications can be inherited from one cell generation to the next and can also be influenced by environmental factors. For example, exposure to certain toxins or malnutrition during prenatal development can lead to epigenetic changes that affect gene expression and increase the risk of certain diseases later in life. Additionally, certain diseases such as cancer are associated with specific abnormal epigenetic modifications.
Understanding the mechanisms of epigenetic regulation and how it affects gene expression is crucial for the development of new therapeutic strategies for diseases. For example, drugs that target enzymes involved in epigenetic modifications have been developed and are being used in clinical trials for the treatment of cancer and other diseases. 🎗️
Gene Expression via Phenotypes
The phenotype of a cell or organism is the observable characteristics of the cell or organism, which is determined by the combination of genes that are expressed and the levels at which they are expressed. 👀
a. Observable cell differentiation results from the expression of genes for tissue-specific proteins. Different cell types in an organism have distinct functions and characteristics, which are determined by the specific set of genes that are expressed in each cell type.
For example, muscle cells express genes for muscle-specific proteins such as actin and myosin, while nerve cells express genes for nerve-specific proteins such as neurofilaments. This process of cell differentiation is tightly regulated by various mechanisms including transcriptional regulation, post-transcriptional regulation and post-translational regulation.
b. Induction of transcription factors during development results in sequential gene expression. During the development of an organism, different sets of genes are expressed at different stages. This sequential gene expression is controlled by a complex network of transcription factors, which are proteins that bind to specific regulatory sequences in the DNA and control the rate of transcription.
These transcription factors are often activated or repressed in a specific temporal and spatial pattern during development, which leads to the expression of the appropriate genes at the appropriate time.
For example, during embryonic development, there are master regulators such as homeobox genes that control the development of different body parts, and the expression of these genes is tightly regulated by a network of other transcription factors. Understanding the mechanisms of gene regulation during development is crucial for understanding the process of development and the causes of developmental disorders.
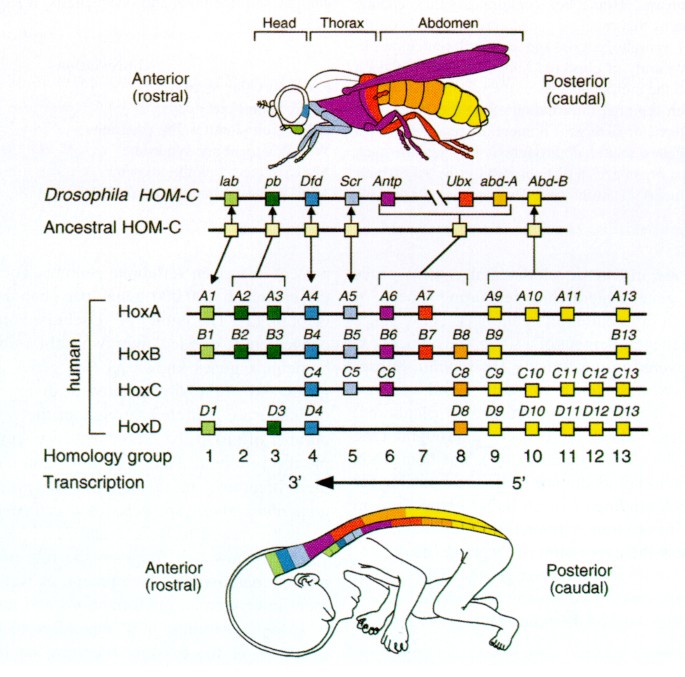
Source: Nature
Coordinated Regulation in Prokaryotes and Eukaryotes
Both prokaryotes and eukaryotes have groups of genes that are coordinately regulated, which means that the expression of these genes is controlled as a group rather than individually. This type of regulation is important for the efficient and coordinated expression of genes that are involved in the same cellular process. 📜
Prokaryotes: Operons
In prokaryotes, groups of genes called operons are transcribed in a single mRNA molecule. These operons are controlled by a single promoter, which is a region of DNA that binds to the RNA polymerase and initiates transcription. An example of an operon is the lac operon, which is responsible for the metabolism of lactose in bacteria. 🦠
The lac operon is an example of an inducible system, which means that the expression of the genes in the operon is increased in the presence of the inducer, lactose. The lac operon is controlled by a repressor protein, which binds to the operator region of the operon and prevents the binding of the RNA polymerase to the promoter. In the presence of lactose, the repressor protein is inactivated, allowing the RNA polymerase to bind to the promoter and initiate transcription.
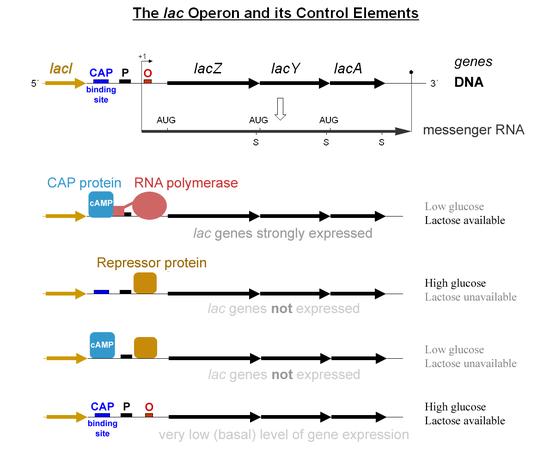
Source: Wikimedia Commons
The operon is regulated by a lac repressor and a catabolic activator protein (CAP). The lac repressor normally will block transcription, but when it senses the presence of lactose, it stops repressing so that the gene can be expressed. The CAP detects glucose, and activates transcription when glucose is low. Both the CAP and the lac repressor detect indirectly. The presence of cAMP lets the CAP know that glucose levels are low, whereas the lac repressor senses the lactose isomer allolactose. lacZ, lacY, and lacA are the genes that are expressed, and can be used for metabolism.
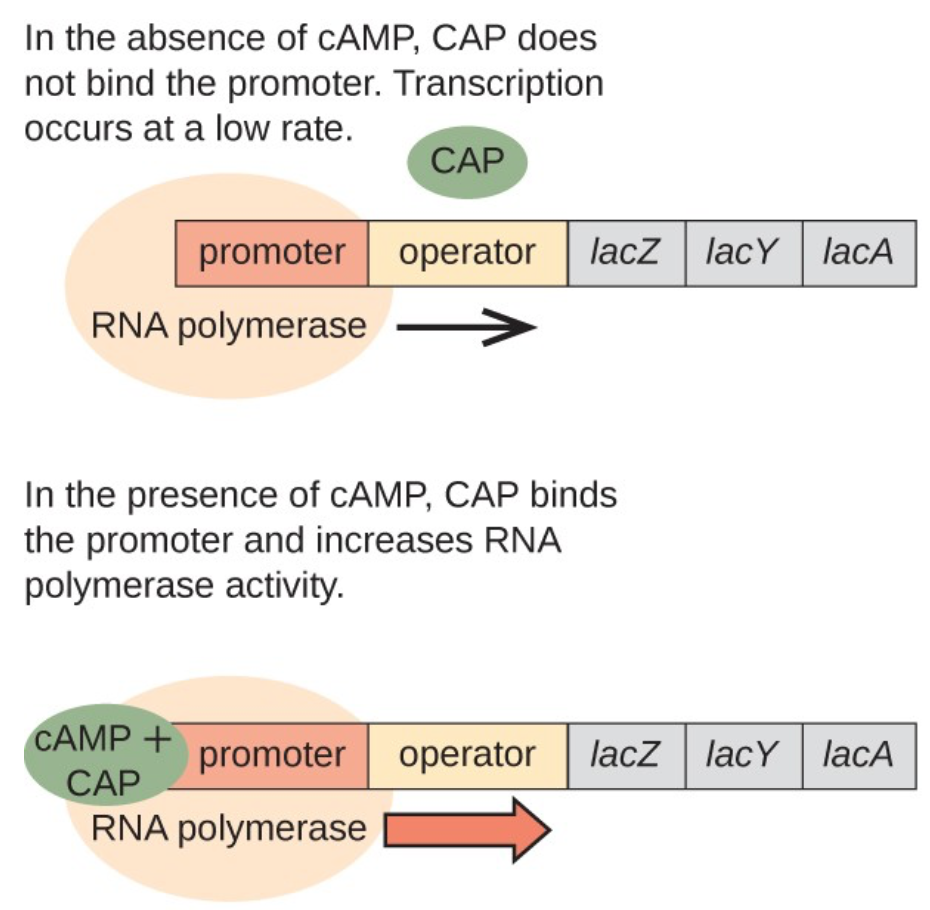
Source: Lumen Learning.
Repressible Operons
Trp operons are also found in E. coli, but they are repressible operons. This means they only turn off when needed. When the repressor is bound to tryptophan, it blocks the expression of the operon. It is only turned off when the trp levels are low. When the repressor is not bound, the operon works very similarly to the lac operon described above. There is a promoter, operator, and various genes that are transcribed as a single mRNA.
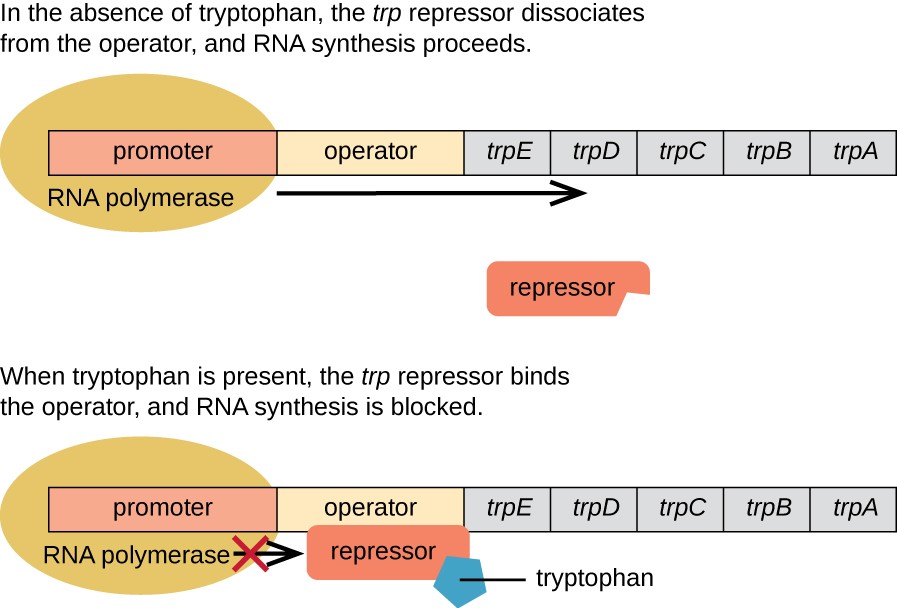
Source: LumenLearning.
Eukaryotes: Similar Transcription Factors
In eukaryotes, groups of genes may be influenced by the same transcription factors to coordinately regulate expression. These transcription factors bind to specific regulatory sequences in the DNA and control the expression of genes.
For example, a group of genes that are involved in cell growth and division may be controlled by the same transcription factor, which would result in the coordinated expression of these genes. Similarly, a group of genes that are involved in the response to a specific environmental stimulus may be controlled by the same transcription factor, which would result in the coordinated expression of these genes in response to the stimulus.
Understanding the mechanisms of coordinated gene regulation in eukaryotes is important for understanding the regulation of cellular processes such as cell growth and development, and the response to environmental stimuli! 🧚
Browse Study Guides By Unit
🧪Unit 1 – Chemistry of Life
🧬Unit 2 – Cell Structure & Function
🔋Unit 3 – Cellular Energetics
🦠Unit 4 – Cell Communication & Cell Cycle
👪Unit 5 – Heredity
👻Unit 6 – Gene Expression & Regulation
🦍Unit 7 – Natural Selection
🌲Unit 8 – Ecology
📚Study Tools
🧐Exam Skills

Fiveable
Resources
© 2025 Fiveable Inc. All rights reserved.